Development of techniques for measurement, manipulation, and theoretical analysis of mechanical self-organization
2025-2026
B02-5
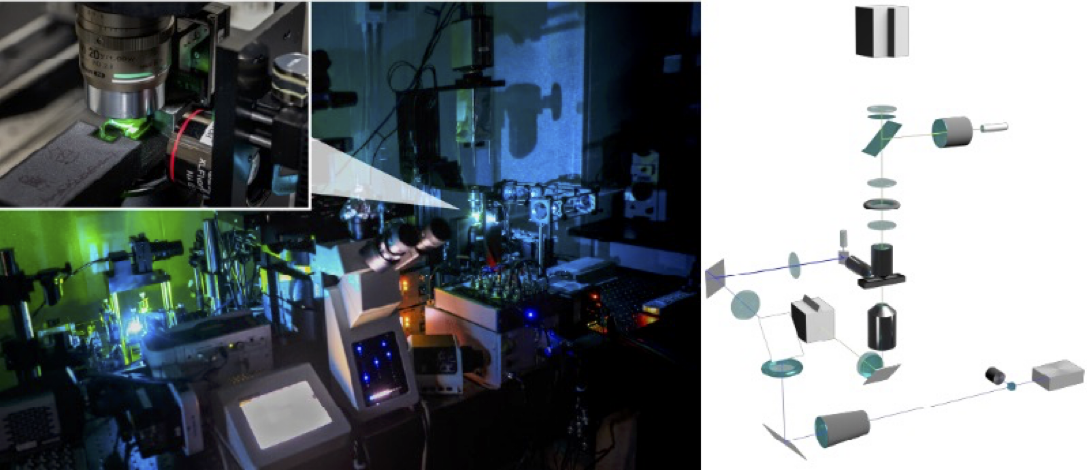
Development of advanced nanoscopy for manipulation of mechanical information within living organisms
Understanding the functions of mechanical information within living organisms requires not only its visualization but also perturbation of the information is necessary. However, due to the technical limitations, manipulating mechanical information at the subcellular level is difficult. In this study, I aim to overcome these challenges by integrating optical tweezers with adaptive optics. This approach achieves a more than a tenfold improvement in precision and sensitivity compared to conventional methods, even within tissues and deep biological environments. The resulting 3D manipulation technique enables non-labeled control of mechanical information at the organelle level within tissues and living organisms.
B02-6
()
()
B02-7
Three-dimensional parameter inference for epithelial mechanics
Measuring mechanical parameters in tissues, such as the elastic modulus of cell-cell junctions, is essential to decipher the mechanical control of morphogenesis. However, their in vivo measurement is technically challenging. Recently, we reported an image-based statistical approach to estimate the mechanical parameters of epithelial cells (Ogita et al. PLoS Comput. Biol., 2022). Candidate mechanical models are constructed based on force-cell shape correlations obtained from image data. Substitution of the model functions into force-balance equations at the cell vertex leads to an equation with respect to the parameters of the model, by which one can estimate the parameter values using a least-squares method. Supported by this consortium, we further reformulated this parameter inference method using Bayesian statistics, allowing the integration of various types of information (Xin, Ogita et al. J. Theor. Biol., 2024). Over the next two years, we aim to extend the image-based parameter inference to three-dimensional systems and to quantify mechanical parameters in complex tissues and organs. By applying our method to Drosophila tissues and mammalian organoids, we will investigate the relationship between tissue curvature, cell arrangement, and mechanical parameters from both experimental and theoretical perspectives. This will allow us to uncover how apicobasal junction remodeling is driven and how three-dimensional tissue morphogenesis is orchestrated.

2023-2024
B02-1
Understanding the mechanisms of neural crest emergence with neural tube reconstruction in vitro
In this research project, we aim to reconstitute the neural tube in vitro by employing a platform for body axis formation. Subsequently, we will investigate the mechanism of the appearance of Neural Crest (NC) cells during neural tube formation, and we will focus on elucidating how the mechano-chemical feedback guides the differentiation of local NC cells.
B02-2
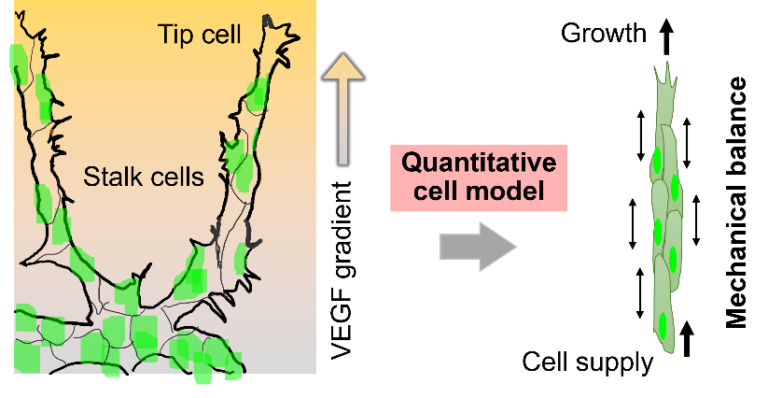
Multicellular mechanics in angiogenesis using quantitative cell motility models
Angiogenesis is an important process in tissue formation and nutrient supply, where cell populations grow while maintaining a one-dimensional shape of tissue. Vascular endothelial growth factor (VEGF) induces angiogenesis, but tip cells do not migrate in the VEGF environment, even when subsequent stalk cells are laser-ablated to isolate the tip cells. It is not yet clear whether tip cells are pulling the stalk cells or if the stalk cells are pushing the tip cells. This project aims to understand the underlying principles of tissue formation during angiogenesis using image analysis and a mathematical model that incorporates mechanistic factors. Since qualitative models have limitations in consistently describing tissue movement according to experimental data, this research project plans to construct a quantitative cell model to reproduce angiogenesis and elucidate the principles from a mechanistic perspective.
B02-3
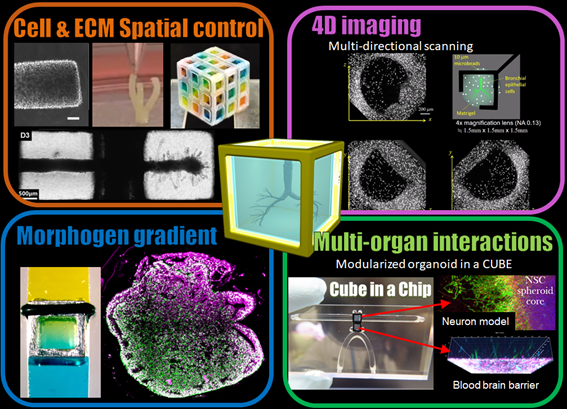
Self-organization of organoids woven by gene expression and mechanical boundary condition
This research aims to develop a method for a combined analysis of the mechanical properties (cell surface tension) and gene expression of individual cells in tissues and organoids. We will explore the temporal variation of gene expression and mechanical properties of cells during organogemesis and tissue remodeling to elucidate the molecular mechanisms that coordinates mechanical self-organization during tissue formation.
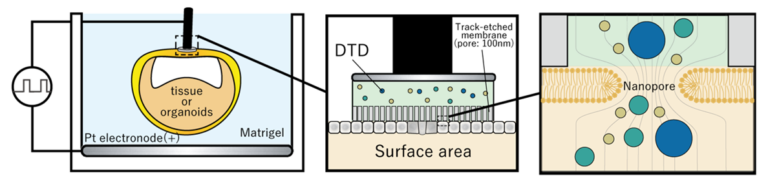
B02-4
Development and application of Bayesian inference of mechanical parameters
Measuring mechanical parameters in tissues, such as the elastic modulus of cell-cell junctions, is essential to decipher the mechanical control of morphogenesis. However, their in vivo measurement is technically challenging. Recently, we reported an image-based statistical approach to estimate the mechanical parameters of epithelial cells (Ogita et al. PLoS Comput. Biol., 2022). Candidate mechanical models are constructed based on force-cell shape correlations obtained from image data. Substitution of the model functions into force-balance equations at the cell vertex leads to an equation with respect to the parameters of the model, by which one can estimate the parameter values using a least-squares method. Over the next two years, we will reformulate our mechanical parameter inference in the framework of Bayesian statistics to incorporate more information such as a group property. We will then apply an extended method to image data that will be acquired in this consortium and we clarify the functional significance of mechanical parameters from both theoretical and experimental perspectives.